Computational Systems Biology

Research Staff
-

Professor
Shigehiko KANAYA -

Affiliate Professor
Hidehiro IIDA -

Affiliate Professor
Tetsuo SATO -

Affiliate Professor
Klaus Walter Lange -

Affiliate Professor
Ming HUANG -

Affiliate Professor
Naoaki ONO -

Associate Professor
Altaf-Ul-Amin -

Assistant Professor
Ahmad Kamal NASUTION -

Affiliate Assistant Professor
Fan ZHANG -

Affiliate Assistant Professor
Ryota WAKAYAMA
Research Areas
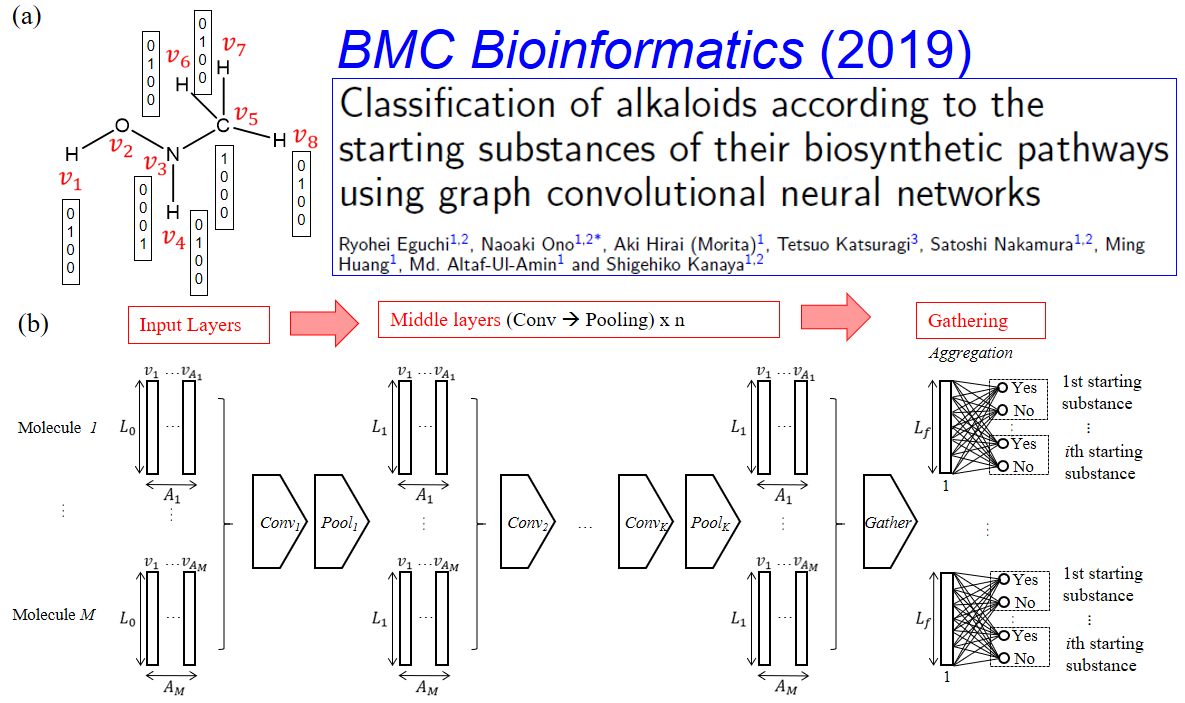
Fig.1 Molecular Graph Convolution Neural Networks (GCNN) makes it possible to predict biological activity and metabolic processes for molecules.
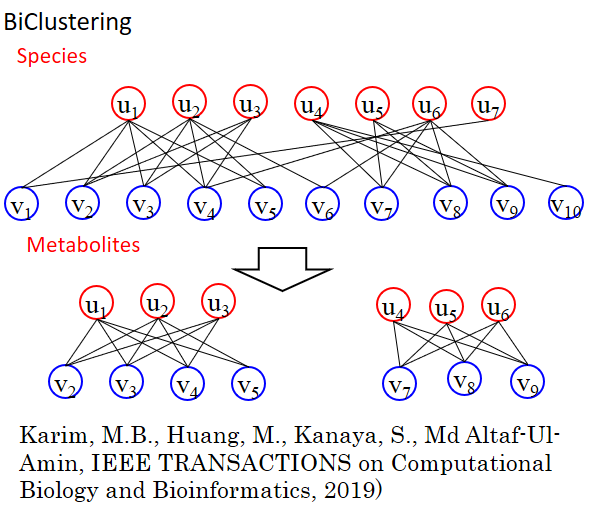
Fig.2 The novel algorithm BiClustering (BiClus) has been developed in our lab, which makes it possible to create groups based on two different attributes.
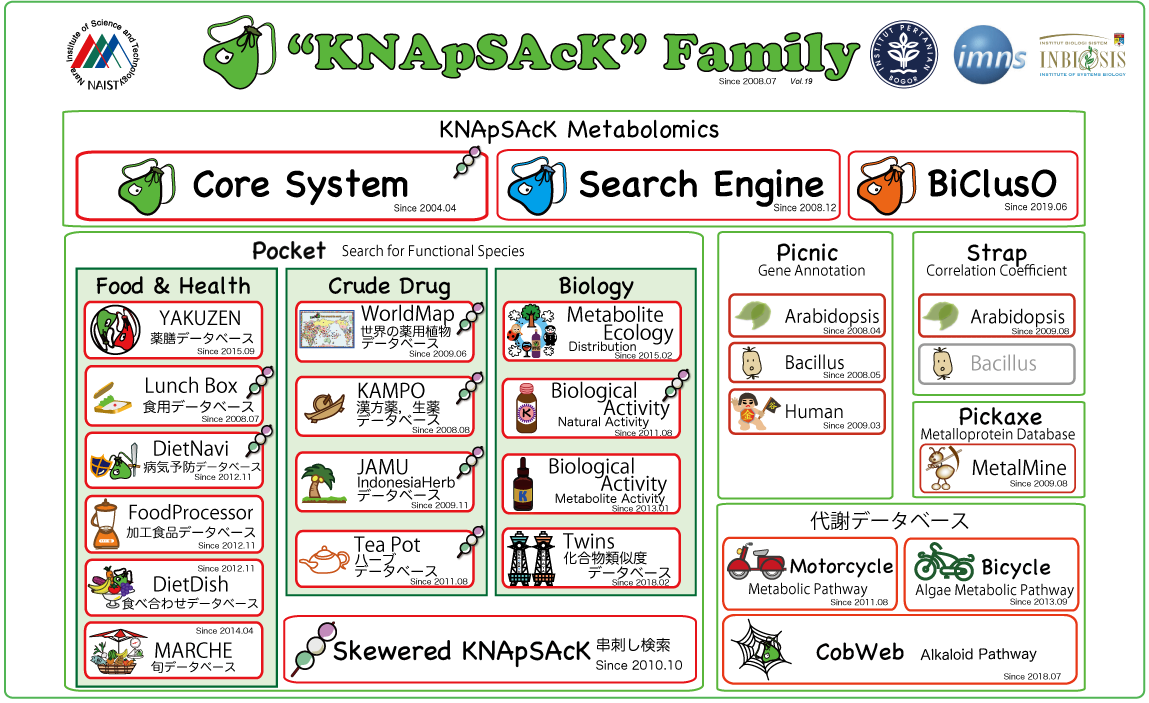
Fig.3 Main page of "KNApSAcK Family DB
(http://kanaya.naist.jp/KNApSAcK_Family/).
This DB has become a world standard database consisting of metabolite-species relationships and cited by very large number of scientific papers.
Biomedical & health data sciencen
In collaboration with medical hospitals and other academic institutions, we are developing various biomedical engineering technologies based on information technology and state-of-the-art deep learning techniques.
By incorporating the strengths of the wearable/unconstrained sensing techniques and information technology such as deep learning techniques (CNN, GAN, etc.), we are developing health monitoring systems for medical and personal use.
- A computer-aided diagnosis assistance system for medical images
- A wearable deep body thermometer monitoring system
- A cuffless blood pressure monitoring system
- A heart health monitoring system based on contactless electrocardiograph
Systems biology & bio data science
Huge biological data sets, such as more than 1,000 genome sequences, have caused a paradigm shift into a holistic approach to understanding living things as systems. In this field, we keep incorporating state-of-the-art data modeling/manipulating techniques such as deep learning techniques to better our understanding.
With the development of omics technologies, it has become imperative to systematically analyze all biological components (genes, mRNA, proteins and metabolites). To meet this challenge, we have developed a clustering algorithm (DPClus, BiClus) to extract highly connected clusters.
Metabolomes & drug discovery
Cells consist of a few thousand molecules. Of those, metabolites are mainly produced by enzymatic reactions. The objective of metabolome analysis is to comprehensively identify which particular metabolites affect cellular networks. As a metabolome analysis platform, we have developed a species-metabolite database, KNApSAcK, covering almost all reported metabolites. To date, 50,048 metabolites and 101,500 species-metabolite relationships have been accumulated. We could extract metabolic pathway information using Molecular Graph Convolution Neural Networks MGCNN).
Key Features
Regardless of research backgrounds, we enjoy an interdisciplinary field between information technology and bio-medical science to mine and integrate knowledge in biology, medical science and health-care. "Let's do research on what we want!" is the motto of our lab.